Structural consequences of Alternative splicing
Alternative splicing
The process of Alternative Splicing (AS) generates alternate spliced mRNAs, which can potentially encode proteins of variable function(s) and thereby contributing to proteome diversity in eukaryotes. Among higher eukaryotes, human genes exhibit alternative splicing in ~95% of multi-exon genes that has been suggested to play an essential role in regulating essential cellular functions such as enzymatic activities, apoptosis, transcription factor activity, apoptosis, autophagy, differentiation, and other developmental processes. Importantly, aberrant or deregulation of AS events have been implicated as cause of several diseases in humans such as cancer, neurological disorders etc. AS derived transcriptome diversity has been extensively characterized using genome-wide microarray and RNAseq analyses. However, the same has not been possible due to technical limitations of in quantitative estimation proteins. Recent advancement in the area of proteogenomics has enabled accurate identification of proteins. With availability of high-throughput MS-MS data from various experimental studies, it is feasible to estimate and identify the tissue/cell level expression of transcripts. Moreover, we can analyze the effect of loss/gain of region of protein in alternatively expressed transcripts on their structure/function. Broadly, in our group we are investigating: - Effect of inclusion/exclusion exons on the structure and function of isoform.
- Understanding the evolution of exons in eukaryotes.
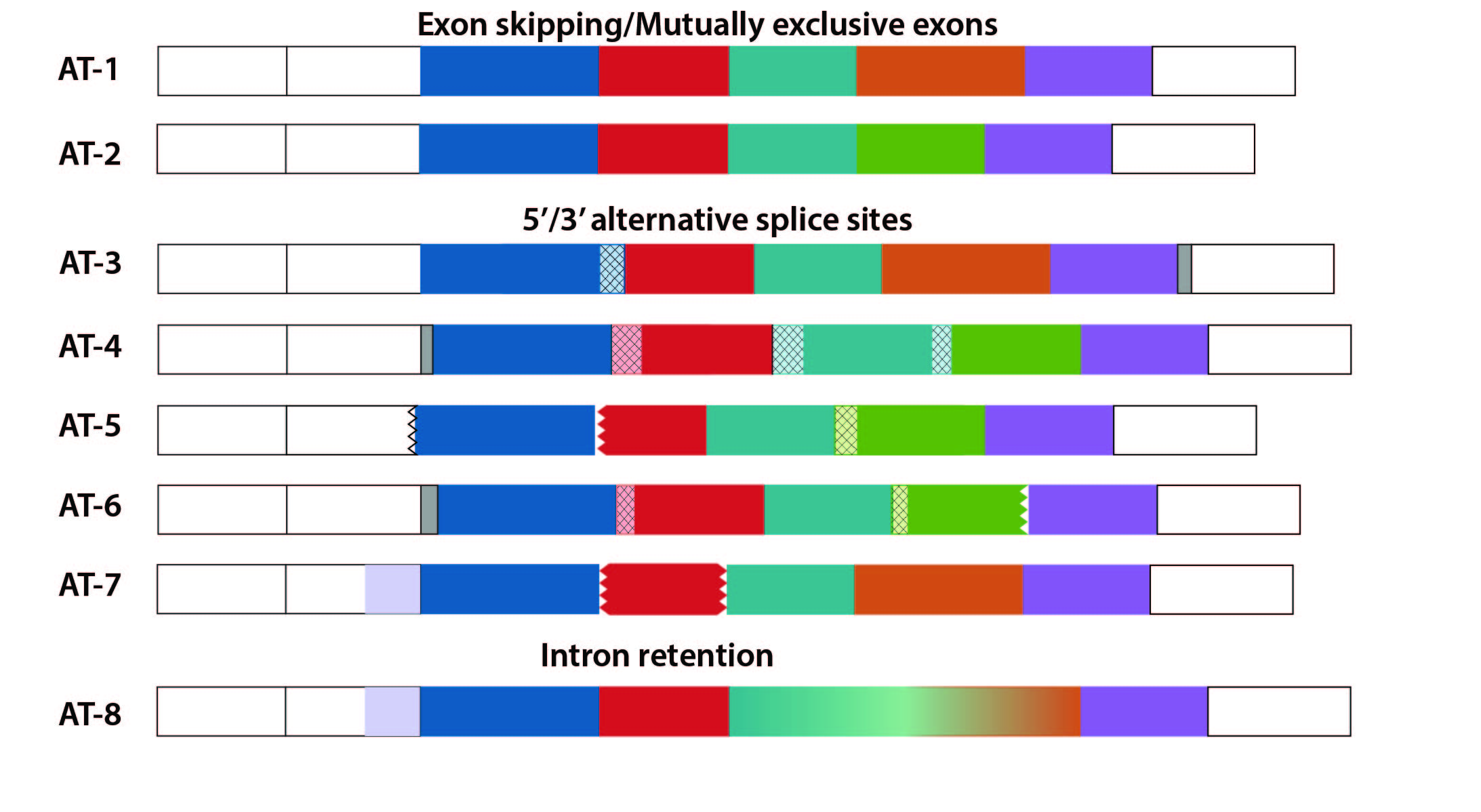
Various Alternative Splicing events
The nomenclature of exon is constructed so that we can reliably study exon combination and its effect on alternatively spliced transcripts or isoforms. The exploration of prevalence of various splicing events and their affect on the transcript(s) or protein sequence in an organism of tracing the evolutionary path or history of exons is both cumbersome and demanding. Moreover, in the era of proteogemonics, an approach to describe transcript(s) through annotated exon(s) can assist in mapping the isoform diversity of a gene. Having employed genomic/transcriptomic features for AS event identifications resulted in complicated exon description. The nomenclature will be explored to study evolution of exons in orthologous proteins across higher eukaryotes.